Fetal RhD Genotyping using Cell Free Fetal DNA from Maternal Plasma in RhD Negative Women in Pakistan
Naeem MA*, Amin N, Javed A, Khan M, Rathore A, Amir M, Mansoor I, Afzal M
Armed Forces Post Graduate Medical Institute Rawalpindi Pakistan
*Corresponding author: Mohammad Abdul Naeem, Armed Forces Post Graduate Medical Institute Rawalpindi, Pakistan
Article History
Received: September 18, 2021 Accepted: September 21, 2021 Published: October 22, 2021
Citation: Naeem MA, Amin N, Javed A, et al. Fetal RhD Genotyping using Cell Free Fetal DNA from Maternal Plasma in RhD Negative Women in Pakistan. Int. J.Obst & Gync. 2021;1(1):16‒19. DOI: 10.51626/ijog.2021.01.00003
Abstract
Background: The Rhesus (Rh) blood group system is one of the most hemogenic and polymorphic system in the body. RhD negative pregnant women carrying RhD positive fetus are always at the risk of hemolytic disease of the fetus and newborn. Prenatal fetal RhD genotyping leads to appropriate management and targeted anti-D immunoprophylaxis in RhD-positive pregnancies only.
Objective: To determine the accuracy and diagnostic feasibility of non-invasive fetal RhD genotyping by analysis of cell free fetal DNA based on real time polymerase chain reaction (RT-PCR).
Materials and Methods: A total of 100 EDTA-blood samples from pregnant women between 10-32 weeks of gestation were collected. The cell free fetal DNA was extracted from maternal plasma followed by PCR analysis for SRY, RhD (exons 5,7,10) and RASSF1A genes using specific primers. Fetus was considered as RhD-positive when amplification of all targeted RhD exons were observed and in case, where no amplification for RhD exons was detected but Beta-globin gene and SRY gene was amplified, fetus were characterized as RhD-negative. When one or two RhD exons were amplified, the results were predicted to be inconclusive and RT-PCR assay with new extracted DNA samples was repeated.
Results: Out of 100 pregnancies, none of the sample was found to be inconclusive for fetal RhD status. No false- positive and false-negative results were registered. The sensitivity, specificity and diagnostic concordance was predicted to be 100%.
Conclusion: The protocol we applied was highly accurate for non-invasive fetal RhD determination. We therefore recommend implementation of non-invasive RhD genotyping in prenatal screening.
Keywords: Cff DNA; Fetal RhD genotyping; Maternal plasma
Introduction
All over the world, Rh incompatibility between RhD-negative mother and RhD-positive fetus is of grave concern due to production of maternal anti-D alloantibodies which is a major cause of haemolytic disease of the fetus and new-born [1-3]. Intervention of anti-D prophylaxis during antenatal and postnatal follow up has decreased the risk of RhD alloimmunization to a greater extent [4]. However, 40% pregnant women receive unnecessarily anti-D immunoglobulin as they carry RhD-negative fetus. Therefore, accurate prediction of prenatal fetal RhD type may assist in the appropriate management of the pregnancy and rationale use of anti-RhD immunoglobulin. Demonstration of prenatal RhD type of the fetus is usually performed by invasive tests including amniocentesis and chronic villus sampling but are associated with increased risk of maternal-fetal haemorrhage and may result in refusal of intervention.
The discovery of circulating cell-free fetal DNA in maternal plasma opened the possibility for prenatal non-invasive diagnostics. The demonstration of chromosome Y signals in pregnant women carrying a male fetus was the first evidence, followed by detection of an RHD gene in RhD negative women. The technique is increasingly used for screening and diagnostics of genetic disorders. Denmark was the first to introduce national prenatal screening or fetal RHD and targeted anti-D prophylaxis in 2010, followed by the Netherlands. 18 In a prospective study of 4118 pregnancies exposed noninvasive fetal RHD screening in the first trimester we found a high sensitivity and specificity for the test.19 Using first trimester screening to target anti-D to RhD-negative women carrying RHD-positive fetuses more than halved the immunisation risk compared with historical comparators receiving postnatal anti-D prophylaxis only.
However, Sweden and other countries have delayed nationwide implementation, awaiting an economic evaluation. Administration of RAADP to all RhD-negative women versus no RAADP has previously been shown to be cost-effective. 1 In publications based on theoretical calculations, fetal RHD screening with targeted antenatal anti-D versus non-targeted RAADP has been judged not to be cost-effective. objective of this study was to investigate the cost-effectiveness targeted antenatal anti-D using fetal Redecreeing in early pregnancy compared with no RAADP or non-targeted RAADP. Therefore, non-invasive diagnostic techniques which are based on extraction of cell-free fetal DNA (cff-DNA) from maternal plasma followed by fetal RhD genotyping has opened new avenues for prenatal diagnosis [5-7].
Materials and Methods
Maternal blood sample collection and plasma DNA extraction
A total of 100 apparently RhD-negative pregnant women who were attending the antenatal care in military hospital, Rawalpindi were purposively included in this study over a period of six months from August 2020 to January 2021. Written informed consent was obtained from each study participant and research protocol was approved by the Institutional Review Board of the Armed Forces Institute of Transfusion Rawalpindi. Their age ranges from 20-35 years and gestational period were spanned between 10-32 weeks. Five millilitres of venous blood sample were collected into K3-EDTA tubes (BD vacutainer, USA) and processed immediately. Plasma was separated by two serial centrifugations, each at 3000g for 10 minutes, and separated supernatant was stored at −40°C in sterile Eppendorf tubes (Axygen, Germany) until further processing. Cell free DNA (cff-DNA) was extracted from maternal plasma using MAGMAX cell free DNA isolation Kit (ABI, USA) according to the manufacturer’s protocol. Presence of β-globin gene was tested as a reference gene to confirm the presence of cff-DNA and SRY gene (male fetal DNA marker) as an internal control to prove the presence of cff-DNA.
Real time PCR
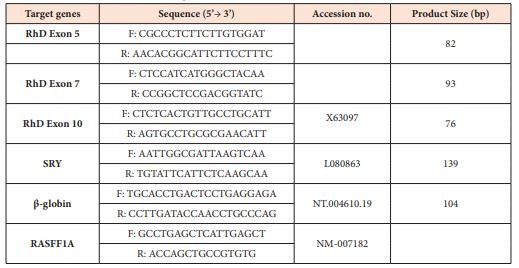
Determination of fetal sex and RhD genotype was carried out by Applied Biosystem ABI 7500 Real-Time PCR System (Thermo Fisher Scientific, USA) using Eva Green Master Mix (2x Maxima Real time PCR, South Korea). Sequences of primers (Macrogen, South Korea) used in the current protocol is given in the Table 1. The isolated cff-DNA was analysed for the presence/absence of RhD gene (exons 5,7,10). The RT-PCR conditions were as follows; two step holding temperatures, 52°C for 2 minutes, 95°C for 10 minutes followed by 50 cycles of 94°C for 45 seconds, 55°C for 45 seconds and 72°C for 45 seconds. Any sample found negative for SRY gene amplification, was predicted to be RhD -negative female fetus. In order to validate the presence of cff-DNA in maternal plasma, an additional test comprising amplification of the hypermethylated RASSF1A gene was performed. A methylation sensitive restriction enzyme (MSRE) BstU1 (Thermo Scientific BSH 1236, USA) was used to digest the predicated RhD/SRY negative samples and after incubation at 60°C for 2 hours, subsequently followed by adding to RT-PCR reactions. Each RT-PCR reaction was performed in duplicate.
Table 1: Sequences of Primers used in the Real Time PCR assay.
Criteria for interpretation of results
Fetus was considered as RhD-positive when amplification of all targeted RhD exons were observed and in case, where no amplification for RhD exons was detected but Beta-globin gene and SRY gene was amplified, fetus were characterized as RhD-negative. When one or two RhD exons were amplified, the results were predicted to be inconclusive and RT-PCR assay with new extracted DNA samples was repeated. Targeted SRY and RASFF1A gene amplification was also considered for determination of male and female fetus respectively. Amplification plots for each targeted gene were analysed and acceptable Cycle threshold values (Ct) were lower than 38.
Postnatal confirmation of the fetal RhD status and sex
All analyses consisted of prenatal RhD genotyping and postnatal RhD phenotyping were carried out blind. Serological testing using anti-D antisera were performed on cord blood samples and neonatal sex was confirmed at birth. Quality Control Positive, negative and blank were run with each PCR reaction.
Statistical analysis
Data analysis was performed using statistical package for social sciences software version 23.0 (IBM SPSS, Chicago, USA). Qualitative variables were presented as percentage or frequency while quantitative variables were presented as mean (SD). The diagnostic concordance was obtained using Cohen’s Kappa. Specificity and sensitivity were calculated using RhD phenotyping and neonate sex as two gold standard method. A total of 100 RhD negative pregnant women were included in the current study. The mean age was 25.4 (3.2) years and a median gestational age was 25 weeks.
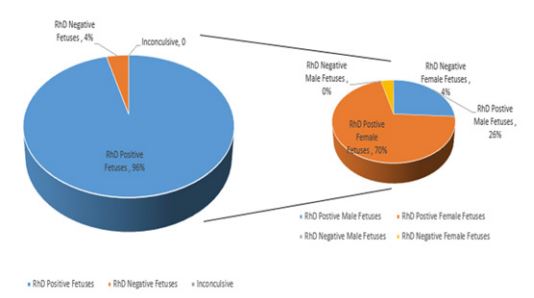
Out of 100 pregnancies, 96.0% RhD positive and 4.0% RhD negative fetus were predicted. Regarding prenatal fetal gender, presence of SRY gene was detected in 26.0% cases characterizing these as male fetuses. Among RhD positive fetus proportion of female fetus was higher whereas no male fetus showed RhD negative blood group (Figure 1). In 74.0% SRY negative cases, cff-DNA was found in all cases using fetal methylation marker which were subsequently characterized as female fetus.
Figure 1: Distribution of male and female fetuses with respect to predicted RhD status.
Prenatal prediction of fetal RhD type and gender from maternal plasma showed 100% accuracy when compared to postnatal serological testing and neonatal sex confirmation respectively. No false-positive or false-negative results were found. Hence, sensitivity, specificity and diagnostic concordance for RhD and SRY genotyping was predicted to be 100%
Discussion
Valuable genetic information can be retrieved using circulating DNA in body fluids. The discovery of cff-DNA has revolutionized the prenatal care by potentially employing cff-DNA in predicting whether a fetus is antigen positive in RhD-negative pregnant women thereby decreasing RhD incompatibility consequences [8]. Fetal RhD genotyping is a routine antenatal testing in developed countries utilizing qPCR as a method of choice with high sensitivity and specificity [9]. However, in developing countries like Pakistan, unavailability of prenatal fetal RhD genotyping is not only a growing concern owing to irrational administration of anti-D prophylaxis in non-isoimmunised mother with RhD-negative fetus but also associated with colossal burden on health care system. This serves as a research gap to set up a protocol for predicting fetal RhD blood group. The present study was aimed at establishing non-invasive prenatal testing (NIPT) protocol using real time PCR assay for simultaneous detection of fetal RhD status and sex. Regarding genetic variations in the Rh blood group system, this may hamper the correct analysis of phenotype. Therefore, the possibility of false-negative and false-positive results cannot be overlooked.
In developing NIPT, selection of RhD genes for qPCR and ascertain the presence of cf-and cff-DNA is inevitable [10]. The current protocol investigated all cf-DNA samples for presence of RhD exons 5,7 and 10. The notion was to increase the sensitivity of the test. RhD exon 5 was selected to distinguish between RhD and RhD pseudogene RHDΨ. Circulating DNA in maternal blood sample which constitutes a total DNA is comprised of maternal and fetal DNA. These two were discriminated on the basis of β-globin and SRY gene sequences which eventually validated the presence of cff-DNA respectively in our study. However, there also exist a possibility that mother may carry female fetus and employing SRY marker in these cases would not be appropriate. Hence, in order to prove the presence of cff-DNA, RASFF1A gene marker was used because this is hypermethylated in fetal DNA and hypomethylated in maternal DNA. After application of the validated technique, we found conclusive results for all 100 investigated pregnancies with 100% accuracy, concordance, sensitivity and specificity. Similar studies were conducted in other Asian countries including Iran and Turkey. In developing countries, the accessibility of current proposed technique is considerable. In our opinion, expensive anti-D immunoprophylaxis and limited availability may enhance the possibility of implementing cost-effective RT-PCR assay for prenatal fetal RhD genotyping using cff-DNA in clinical practice [11].
Cell-Free Fetal DNA Enrichment
Now a days DNA enrichment is performed after end-repairing and before adaptor ligation and NIPS library constructed. Magnetic beads with an average particle size of 1μm are used for the purpose of size-selecting the end repaired DNA fragments with size smaller than 160 bp. To achieve the highest efficiency, the optimize step is taken by testing a series of different bead concentrations. Magnetic beads are added to the end-repaired DNA fragments, followed by vibrating the tubes for at least 3 s. The tubes are then suspended for 5 min and transferred to a magnetic rack. The supernatant containing size-selected DNA fragments is then transferred to another tube for adaptor ligation. This latest technique has changed the horizons of fetal DNA concentration process.
Conclusion
To conclude, our study demonstrated 100% accuracy for prenatal fetal RHD genotyping using cff-DNA circulating in maternal plasma. The non-invasive testing using RT-PCR assay predicts fetal RhD status with 100% sensitivity and specificity and would assist in preventing anti-D immunoprophylaxis in RhD negative women carrying RhD negative fetus. Our study demonstrated small proportion of RhD negative fetus 4.0% but conducting similar study with larger sample size, could escalate this number in future. Therefore, we recommend to include highly accurate non-invasive prenatal testing as an obligatory part in clinical laboratory routine in Pakistan which also leads to timely management of pregnant women bearing RhD positive fetus.
References
- Hamilton PJ (1998) Blood Transfusion in Clinical Medicine. Journal of Clinical Pathology 51: 640.
- Loy YD, Hjelm NM, Fidler C, Sargent IL, Murphy MF, et al. (1998) Prenatal diagnosis of fetal RhD status by molecular analysis of maternal plasma. New England Journal of Medicine 339(24): 1734-1738.
- Daniels G, Finning K, Martin P, Massey E (2009) Noninvasive prenatal diagnosis of fetal blood group phenotypes: current practice and future prospects. Prenatal Diagnosis 29(2): 101-107.
- Perry GH, Xue Y, Smith RS, Meyer WK, Caliskan M, et al. (2012) Evolutionary genetics of the human Rh blood group system. Hum Genet 131(7): 1205-1216.
- Bombard AT, Akolekar R, Farkas DH, VanAgtmael AL, Aquino F, et al. (2011) Fetal RHD genotype detection from circulating cell-free fetal DNA in maternal plasma in non-sensitized RhD negative women. Prenat Diagns 31(8): 802-808.
- Swanson A, Sehnert AJ, Bhatt S (2013) Non-invasive prenatal testing: technologies, clinical assays and implementation strategies for women’s healthcare practitioners. Curr Genet Med Rep 1(2): 113-121.
- Kolialexi A, Tounta G, Mavrou A (2010) Noninvasive fetal RhD genotyping from maternal blood. Expert Rev Mol Diagn 10(3): 285-296.
- Lo YM, Hjelm NM, Fidler C, Sargent IL, Murphy MF, et al. (1998) Prenatal diagnosis of fetal RhD status by molecular analysis of maternal plasma. N Engl J Med 339(24):1734-1738.
- Akolekar R, Finning K, Kuppusamy R, Daniels G, Nicolaides KH (2011) Fetal RHD Genotyping in Maternal Plasma at 11-13 Weeks of Gestation. Fetal Diagnosis and Therapy 29: 301-306.
- Liao GJ, Chiu RW, Lo YM (2012) Prenatal assessment of fetal chromosomal and genetic disorders through maternal plasma DNA analysis. Pathology 44(2): 69-72.
- Ping H, Dong L, Yangyi C, Lin Y, Qiao F, et al. (2019) An enrichment method to increase cell‑free fetal DNA fraction and significantly reduce false negatives and test failures for non‑invasive prenatal screening: a feasibility study. J Transl Med 17(1): 124-127.